Section 10 - Data Processing
This section covers several topics including: Data Ingestion, HIVE / CODCC Validation, and Processing. You can also see the flow of data uploads and datasets through the system as well and more information about the various stages/statuses that occur during these processes.
Data uploads go through several statuses during the ingest process.
See a Data Ingest Flow Diagram
#### Data Upload Flow Diagram 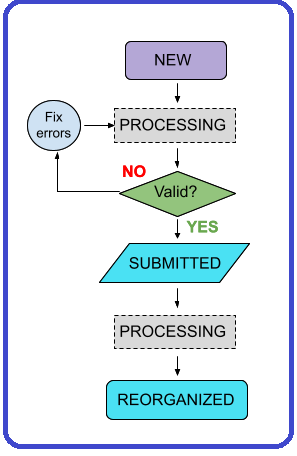See details about Ingestion Statuses
### Ingestion Statuses These statuses display in the ingestion portal as a data set is processed. **New** - Data upload registered, Globus upload directory created. Data provider has uploaded data. After HIVE (or CODCC) validation, status changes to Submitted when the data provider hits the submit button on the upload. Prerequisite: Local validation of data by provider prior to data upload. **Processing** - The data upload is being processed and is not editable. A transient state (between other states) while automated processes act on the upload. **Valid** - Every data upload is reorganized into data sets. This is a semi-automated process. If the data upload is valid, data curation can kick off this process. **Submitted** - Data upload submitted for validation and processing by the HIVE or CODCC. The data upload can be automatically or manually ingested. Status changes to _Processing_ when data curation presses the “Validate” button. **Reorganized** - Data curation hits the “Reorganize” button to kick off automated processing that generates the data sets. The status of the upload changes to _Reorganized_ when this completes. **Invalid** - The data upload did not pass HIVE (or CODCC) validation or a failure occurred during processing. Someone from the HIVE (or CODCC) will contact the data submitter to address this status. **Error** - An (unspecified) error occurred during HIVE or CODCC processing.
Data Processing: Ingestion, HIVE / CODCC Validation, and Processing
Prerequisites
- Upload all data files to the team’s Globus folder.
- Each dataset has a corresponding directory and row in an associated metadata template file.
- Each row in a template = one dataset
- Multiple metadata template files can be included, but at least one is required.
NOTE: Multiple sets of data can be uploaded and the same data set could be uploaded more than once. Each data upload is assigned a UUID (universally unique identifier), but not a version number, until it has completed the entire data ingest, validation, and approval process and is published.
-
Email the HuBMAP Helpdesk OR SenNet Helpdesk once all the files associated with an upload have been uploaded. This lets them know that the upload is ready for ingestion (not an automated process).
IMPORTANT: Include the root path(s) of the specific data upload(s) in the email.
- The HIVE (or CODCC) extracts: Each data upload, corresponding data, and registers them as individual datasets.
-
The HIVE (or CODCC) processes: The data and metadata that you have uploaded for ingestion. If ingestion fails, or if additional information is needed, the data provider will be contacted using the ticketing system.
NOTE: For Clinical assay data, the Pitt team will de-identify this data and submit the scrubbed data to the HIVE / CODCC. The provider of the data will need to review and approve the release of the de-identified data in the publish step.
- Pipeline processing: The HIVE / CODCC processes certain assays by standardized pipelines (where applicable).
- CODEX - Cyclic immunofluorescence imaging: The pipeline uses Cytokit + SPRM
- ATAC-seq - (Including sc-, sn-, and bulk variants): The pipeline uses SnapTools + SnapATAC
- RNA-seq - (Including sc-,sn-,bulk): The pipeline uses Salmon + Scanpy + scVelo
- Other imaging data: SPRM
Additional pipelines will be added over time, as needed. Pipelines are available for download use by others, including TMCs.
NOTE: Generally, TMCs are not involved with pipelines, but may be contacted if an error occurs.