Example Data Query and Download
Overview
The combination of the RESTful parameterized search and the SenNet Command Line Transfer Tool provides for an easy way to programmatically query SenNet data and download the results of the query.
Description
Below is an example of how to use the RESTful parameterized search endpoint to query for datasets with specific attributes and produce a manifest of datasets to download and how to use the manifest to download all of the data for the referenced Datasets. The parameterized search feature shown in this example is a simple query mechanism that allows quick querying of data via a single RESTful URL call where queried attributes are constrained to exact string matches of a limited set of attributes, where the query is an “AND” filtered query with all attribute matches as terms in the “AND” clause, for example the query /param-search/datasets?status=Published&dataset_type=CODEX will return all datasets that are “Published AND a result of a CODEX assay”. If more complex queries are desired use the standard /search endpoint which is documented in the SenNet Search API Endpoints.
This example uses the command line tool curl to execute queries. The Example Data Query and Download Jupyter Notebook has this same example using Python.
Example Query and Download
The following query will return all Histology (dataset_type=Histology) Datasets run on a Keyence BZ-X800 machine (metadata.acquisition_instrument_model=BZ-X800) where tissue from a heart was used (origin_samples.organ=UBERON:0000948). See the RESTful parameterized search page for further information on querying dataset, organ (origin_samples.organ represents the organ in the query and UBERON:0000948 is the UBERON organ code (organ code list available here) and dataset metadata fields.
GET https://search.api.sennetconsortium.org/param-search/datasets?dataset_type=Histology&metadata.acquisition_instrument_model=BZ-X800&origin_samples.organ=UBERON:0000948
As is, if this query is submitted via HTTP GET it will produce a json Response with an array of dataset objects which match the query. Adding the produce-clt-manifest=true option to this query will instead produce a list of Dataset IDs pointing to the Datasets that match this query in a format that will be directly usable by the SenNet Command Line Transfer Tool.
To run this from the command line and save the results to a file run:
curl "https://search.api.sennetconsortium.org/param-search/datasets?dataset_type=Histology&metadata.acquisition_instrument_model=BZ-X800&origin_samples.organ=UBERON:0000948&produce-clt-manifest=true" > dataset-manifest-for-download.out
This results in a file that looks like:
SNT948.QRZW.946 /
SNT976.WLTL.469 /
...
To use the SenNet CLT tool to download the data from these datasets:
- Install the Globus Connect Personal client and the SenNet CLT per the SenNet CLT Setup Instructions
- Python 3.9 or greater is required for the SenNet CLT, install from the Python Downloads page
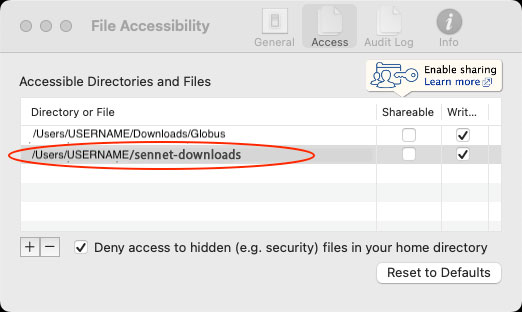
- Setup Note: A common issue arises between the configuration of the GCP client and SenNet CLT. By default, SenNet CLT stores files in the user’s home directory under a directory called
sennet-downloads, so make sure to configure the GCP client by going to “Preferences”–>”Access” and adding thesennet-downloadsdirectory in the user’s home like (Example shown is Mac OS X):
- On the command line log into the SenNet Globus server using:
sennet-clt loginGlobus login screen will open in your default web browser. Follow the instructions to log in. For publicly available SenNet data any login will work (your institution, Google, GitHub, etc..).
- Download the data using the manifest file generated above:
sennet-clt transfer dataset-manifest-for-download.out
Futher instructions on the usage of the SenNet CLT are available on the main SenNet Command Line Transfer Tool page